Introduction
Today, we’re introducing Project OSSAS in collaboration with LAION and Wynd Labs. Project OSSAS is a large-scale open-science initiative to make the world’s scientific knowledge accessible through structured, AI-generated summaries of research papers. Built on the foundation of Project Alexandria, OSSAS uses custom-trained Large Language Models, and idle compute sourced from tens of thousands of computers around the world to process scientific research papers into a standardized format. This machine-readable format can be explored, searched, and linked across scientific disciplines.
Contributors: Christoph Schuhmann, Amarjot Singh, Andrii Prolorenzo, Andrej Radonjic, Sean Smith, and Sam Hogan
Models: OSSAS-Qwen3-14B, OSSAS-Nemotron-12B
Visualizer: https://ossas.inference.net
Visualizer Code: View on GitHub
Paper: Coming soon
The work to complete this project also spans many disciplines: bespoke model-training pipelines, high-throughput distributed-systems for fetching data and processing inference on idle compute, cryptographic protocols to ensure integrity, and more. These components are:
Inference Devnet (link) - Galactic-scale distributed networking infrastructure for LLM inference workloads.
OSSAS Models (link) - Nemotron and Qwen models post-trained to process scientific literature at frontier quality and low cost.
Inference Staking (link) - Solana-based staking and emissions protocol to coordinate daily payouts of USDC (write up coming soon)
LOGIC (link)- LogProb based inference verification to enforce honesty amongst network participants.
In this post, we explore Project OSSAS from a high-level and dive deeper into the process we went through to train, evaluate, and serve the custom language models that provide the backbone of the initiative.
Background
Access to scientific knowledge remains one of the great inequities of our time. Paywalls, licensing restrictions, and copyright constraints create barriers that slow research, limit education, and concentrate knowledge in institutions wealthy enough to afford access. A researcher in a developing country, a citizen scientist pursuing independent work, or students at under-resourced universities often cannot access the papers they need.
Additionally, the sheer volume of scientific literature makes it nearly impossible to keep up. Millions of papers are published each year across thousands of journals and preprint servers. Different papers use different formats, structures, and writing styles. Comparing claims across papers, understanding methodological variations, or identifying contradictions requires painstaking manual work.
The goal of Project OSSAS is to transform this landscape by creating a standardized structure for generating summaries of scientific papers at scale using custom-trained Large Language Models.
Project Alexandra established the legal and technical foundation for this work, demonstrating that we can extract factual knowledge from scholarly texts while respecting copyright through "Knowledge Units": structured representations that preserve scientific content without reproducing copyrighted expression.
Building on that foundation, we've spent the last four months developing a complete pipeline for processing scientific literature at scale. Today, we're releasing the first preview of our progress. This includes:
- OSSAS Models: fine-tuned Qwen 3 14B and Nemotron 12B models for structured summarization
- An initial batch of 100,000 research paper summaries in standardized JSON format
- Interactive dataset exploration at https://ossas.inference.net
- Complete evaluation framework and training approach
Our fine-tuned models achieve performance comparable to leading closed-source models like GPT-5 and Claude 4.5 Sonnet at 98% lower cost, while offering the transparency and accessibility that open science demands. This represents the first step toward our larger goal: processing and structuring knowledge from 100 million research papers.
100 Million Papers to Structured Knowledge
Our primary dataset consists of approximately 100 million research papers retrieved from the public internet through a collaboration with Wynd Labs using Grass network. We deduplicated this massive collection and supplemented it with papers from established sources:
- bethgelab's paper_parsed_jsons dataset: High-quality parsed research papers
- LAION's COREX-18text dataset: Diverse scientific texts
- PubMed articles from Common Pile: Biomedical research literature
- LAION's Pes2oX-fulltext dataset: Full-text scientific articles
This represents one of the largest collections of scientific literature ever assembled for knowledge extraction. The scale is crucial to maximize coverage across disciplines, methodologies, and decades of research.
For model training, we created a curated subset of 110,000 papers, carefully sampled to ensure diversity across sources and scientific domains. This subset was divided into 100,000 training samples and 10,000 validation samples. The papers are substantial, averaging 81,334 characters each, with a median of 45,025 characters.
Designing the Schema
We developed a comprehensive Json schema for structuring scientific paper summaries, inspired by the Alexandria project's Knowledge Units concept. The schema classifies articles into three categories:
SCIENTIFIC_TEXT: Complete scientific research articles
PARTIAL_SCIENTIFIC_TEXT: Partial scientific content
NON_SCIENTIFIC_TEXT: Non-research content
For scientific texts, the schema extracts detailed information including title, authors, publication year, research context, methodology, results, claims, and other critical elements. The full schema is available at the bottom of this article.
This structured format makes papers directly comparable and enables novel ways to explore scientific literature. Instead of searching through unstructured text, researchers can query specific fields to find all papers using a particular methodology, compare results across studies, or identify contradictory claims in the literature.
Model Training
To generate these structured summaries at scale, we fine-tuned two open-source models:
Qwen 3 14B: Alibaba's dense transformer model, known for strong reasoning capabilitiesNemotron 12B: NVIDIA's hybrid Mamba-Transformer architecture, offering high throughput
Both models were post-trained on our curated 110,000-paper dataset using summaries generated from top closed-source models as training targets. The training transforms general-purpose open source models into specialized scientific summarizers that understand the conventions, structure, and nuances of research papers across diverse domains.
Validating Quality
For a project of this ambition, evaluation is critical. We can't just claim our models work—we need to demonstrate it through multiple complementary approaches.
LLM-as-a-Judge Evaluation
Our primary evaluation uses an ensemble of three frontier models (GPT-5, Gemini 2.5 Pro, and Claude 4.5 Sonnet) to rate summaries on a 1-5 scale. This multi-model ensemble reduces bias from any single judge and provides robust assessment across multiple dimensions: factual accuracy, completeness, structural adherence, and clarity.
The results validate that well-tuned open-source models can match closed-source performance:
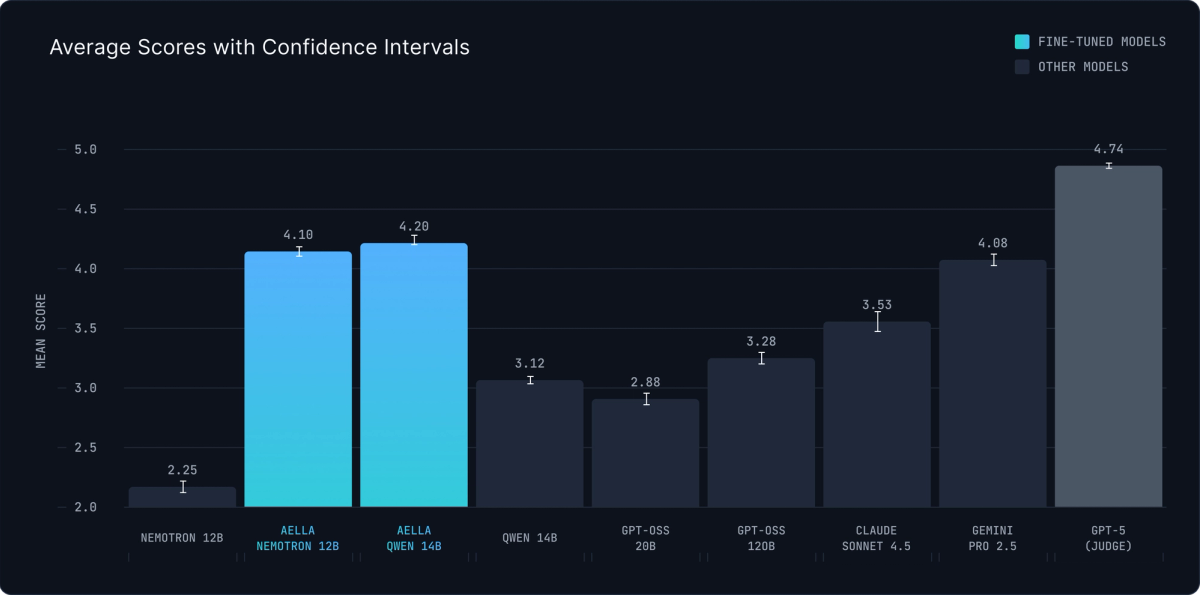
Our fine-tuned models achieve scores within 15% of GPT-5 itself—remarkable performance considering they're open-source, smaller, and far more efficient to run. The substantial gap between base and fine-tuned versions demonstrates the value of domain-specific training for scientific summarization.
QA Dataset Evaluation: Measuring Factual Utility
To validate factual accuracy and downstream utility, we created a complementary evaluation inspired by Alexandria's approach. For a holdout set of papers, we generated 5 multiple-choice questions per paper using GPT-5, then tested whether models could answer these questions based solely on their generated summaries (truncated to 10,000 characters).
This tests something crucial: can the summaries actually support downstream reasoning and information retrieval?
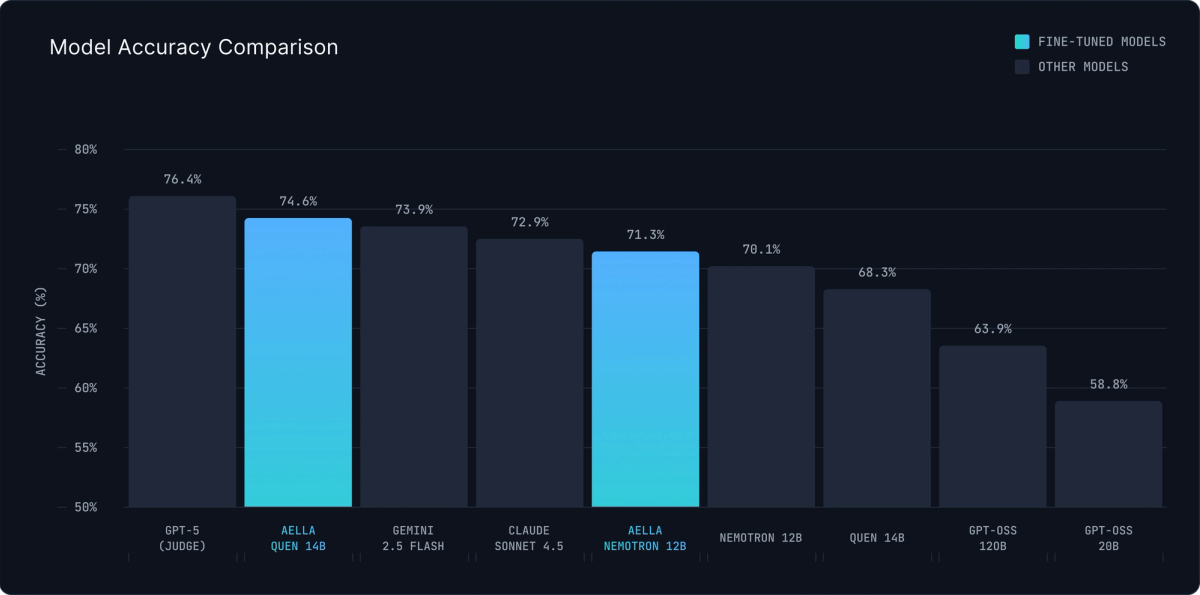
Our models match or exceed leading closed-source alternatives. The Qwen 3 14B fine-tuned model ties with Gemini 2.5 Flash at 73.9%, falling just short of GPT-5's 74.6% score.
Together, these two evaluation methods provide robust validation: the LLM-as-a-judge assessment confirms our summaries are high-quality and well-structured, while the QA dataset proves they're factually grounded and useful for downstream tasks.
Economics and Throughput
Performance on benchmarks matters, but so does practical deployability. Can we feasibly process 100 million papers with these models? How much will it cost in GPU hours to process the entire dataset?
We measured throughput on an 8×H200 node using vLLM:

The OSSAS Nemotron 12B model achieves 2.25x higher throughput than OSSAS Qwen 3 14B across all metrics, including requests per second, input token processing, output generation, and single-request decode speed. For large-scale processing, this difference translates directly to a 2.25x lower total compute cost.
The economic comparison is striking:
- Processing 100M papers with GPT-5: Over $5 million at current pricing
- Processing with our custom models : Under $100,000
This is a 50x cost reduction, all while maintaining comparable quality. Without such a sharp drop in cost, it is unlikely we would be able to fund the entirety of this project . Using custom LLMs, we were able to make scientific knowledge extraction feasible at a scale that was previously prohibitive for all but the wealthiest organizations in the world.
Idle Compute: Infrastructure for Open Science
Even with the substantially lower cost of fine-tuned models, processing 100 million papers requires enormous computational resources. Traditional cloud computing would make this prohibitively expensive. We offer an alternative: Inference Devnet – a permissionless decentralized GPU network that enables crowdsourcing of idle compute resources globally. You can think about Inference Devnet as a modern version of SETI@Home, purpose-built for processing large-scale LLM workloads. Inference Devnet provides massive computational capacity through a network of distributed GPUs running in contributor’s homes and data centers around the world. We believe idle compute holds the key to unlocking the next step in scaling computational resources for scientific research, allowing individuals to contribute their idle resources to scientific initiatives they find compelling, while simultaneously giving underresourced scientists a path to pushing their research forward in a meaningful way. Stay tuned; we will be sharing more about this soon.
Visualization and Discovery
To demonstrate the utility of structured summaries, we built an interactive visualization tool using our first 100,000 summaries.

The visualizer creates a physical representation of the scientific literature using UMAP clustering on embeddings generated from the summaries (using Qwen 3 Embedding 4B). Papers cluster by topic, methodology, and domain, revealing the landscape of scientific knowledge in intuitive visual form.
Key features:
- Spatial clustering: Related papers appear near each other
- Similarity search: Find papers using cosine similarity on summary embeddings
- Interactive exploration: Click papers to see their structured summaries
- Relationship discovery: Identify connections and patterns across the literature
This is just one example of what becomes possible with structured summaries at scale. Once scientific knowledge is in a standardized format, we can build tools for comparative analysis, claim verification, contradiction detection, knowledge graph construction, and other applications that are impractical with unstructured text.
The visualization demonstrates that structured summaries aren't just abstractions; they enable new ways of exploring and understanding scientific literature that simply weren't possible before.
Limitations
Hallucination and Factual Precision
Large language models, even frontier ones, can generate fluent but subtly incorrect statements. This risk is particularly acute for fine-grained scientific details: sample sizes, effect sizes, confidence intervals, exact dates, and unit conversions. Our evaluation framework validates that summaries are useful and well-formed, but cannot guarantee line-by-line factual accuracy for every atomic claim.
The QA evaluation demonstrates that summaries support downstream reasoning effectively, but this doesn't certify that every number, citation, and domain-specific detail is perfectly exact.
Context Limits and Coverage
Current context limits (128,000 tokens in our setup) can force selective reading for extremely long documents, potentially omitting details that matter for particular users or tasks. While our average paper length fits comfortably, edge cases exist where important information might be truncated.
Domain Heterogeneity
Scientific fields have vastly different methodological norms. A physics paper, a clinical trial, a qualitative sociology study, and a computer science benchmark paper all require different types of information extraction. Our unified schema serves broad needs well but may reduce recall for highly specialized subdomains without additional domain-specific post-training.
Appropriate Use
Given these considerations, we emphasize appropriate use: our summaries are high-quality overviews that make literature more searchable, comparable, and explorable. They are educational and research aids that accelerate orientation and review.
For high-stakes scientific activities, such as engineering implementations, clinical or policy decisions, writing scholarly articles or theses, users should verify specific details (numbers, dates, specialized terminology) against original papers. The summaries are starting points for exploration, not replacements for careful reading of primary sources when precision matters.
We provide these summaries openly precisely so the community can validate, critique, and improve them.
Scaling to 100 Million Papers
This release of 100,000 summaries represents our first milestone. Here's what comes next:
Processing the Full Corpus
Our immediate priority is processing the complete ~100 million paper collection with our fine-tuned models. Each paper will receive a standardized structured summary, creating the most comprehensive knowledge extraction from scientific literature ever assembled.
Linking to OpenAlex Metadata
We'll join each summary to OpenAlex metadata, enabling users to traverse authors, venues, institutions, concepts, references, and citations. This creates a graph-native exploration layer where summaries are anchored to bibliographic context and citation neighborhoods. You can navigate not just what papers say, but how they connect to the broader research ecosystem.
Releasing Permissive Licensed Full Texts
For papers with permissive licenses (subsets from PeS2o and Common Pile PubMed), we'll release corresponding full texts paired with summaries. This supports long-context training and benchmarking: AI agents can retrieve and justify claims from millions of documents, using summaries as query scaffolds while grounding answers in released full texts.
Converting to Knowledge Units
Following our Alexandria framework, we aim to iteratively convert the corpus into structured, style-agnostic Knowledge Units that capture entities, attributes, and relations without reproducing copyrighted expression. This yields a shareable factual substrate legally and technically suited for open dissemination and machine use.
Knowledge Unit conversion is substantially more compute-intensive than summarization, so our phased approach prioritizes scaled summaries and metadata linkage first, followed by gradual Knowledge Unit conversion.
Each of these steps moves us closer to the goal: making scientific knowledge truly accessible, comparable, and explorable at global scale.
Open Models, Open Science, Open Future
Custom models can effectively generate high-quality structured summaries of scientific papers at scale. Our fine-tuned models achieve performance comparable to leading closed-source alternatives while offering superior economics, transparency, and accessibility.
The release today, 100,000 summaries, two fine-tuned models, and an interactive visualization tool, represents tangible progress toward a larger vision: processing and structuring knowledge from 100 million research papers to make scientific knowledge truly accessible to anyone with curiosity and an internet connection. This is the beginning of open infrastructure for open science.
Authors: Christoph Schuhmann, Amarjot Singh, Andrii Prolorenzo, Andrej Radonjic, Sean Smith, and Sam Hogan
Models: OSSAS-Qwen3-14B, OSSAS-Nemotron-12B
Visualizer: https://ossas.inference.net
Visualizer Code: View on GitHub
Paper: Coming soon
Acknowledgments: This work was made possible through collaboration with Wynd Labs, LAION, and the broader open science community. We thank the contributors to bethgelab, PeS2o, Common Pile, and OpenAlex whose datasets and infrastructure made this research possible.
Own your model. Scale with confidence.
Schedule a call with our research team to learn more about custom training. We'll propose a plan that beats your current SLA and unit cost.